BREAKING Publication: Proximal Origin of Epidemic Highly Pathogenic Avian Influenza H5N1 Clade 2.3.4.4b and Spread by Migratory Waterfowl
Published evidence suggests current outbreak came from a laboratory.
By Peter A. McCullough, MD, MPH, Nicolas Hulscher, MPH, John Leake
For the last four months we have been barraged with news about highly pathogenic avian influenza (H5N1) or bird flu. We have been told that government agencies are going onto poultry and dairy farms and performing mass PCR testing. With positive tests, large numbers of birds are being culled—that is, intentionally destroyed. So far, only three human cases have been reported, all of them mild. Where is the current strain coming from? How is it spreading?
McCullough Foundation investigators— epidemiologist Nicolas Hulscher, investigative author John Leake, and clinician Dr. Peter McCullough—have just published an intensively referenced manuscript based on a meticulous examination of the large, peer-reviewed literature on the new bird flu strain known as Highly Pathogenic Avian Influenza H5N1 Clade 2.3.4.4b.
The following is our Introduction to the paper, which we took pains to write as clearly as possible for a general readership in a style unencumbered by needless technical jargon.
INTRODUCTION
As of May 29, 2024, there has been significant media coverage concerning a new highly pathogenic variant of H5N1 avian influenza, commonly referred to as “bird flu.” According to a May 16, 2024 advisory issued by the World Health Organization:
The goose/Guangdong-lineage of H5N1 avian influenza viruses first emerged in 1996 and has been causing outbreaks in birds since then. Since 2020, a variant of these viruses has led to an unprecedented number of deaths in wild birds and poultry in many countries. First affecting Africa, Asia and Europe, in 2021, the virus spread to North America, and in 2022, to Central and South America. From 2021 to 2022, Europe and North America observed their largest and most extended epidemic of avian influenza with unusual persistence of the virus in wild bird populations. Since 2022, there have been increasing reports of deadly outbreaks among mammals also caused by influenza A(H5) – including influenza A(H5N1) – viruses. There are likely to be more outbreaks that have not been detected or reported. Both land and sea mammals have been affected, including outbreaks in farmed fur animals, seals, sea lions, and detections in other wild and domestic animals such as foxes, bears, otters, raccoons, cats, dogs, cows, goats and others.
The variant referred to in this WHO advisory is known as highly pathogenic avian influenza (HPAI) H5N1 clade 2.3.4.4b. A literature review on this variant revealed that it—and the subtype (H5Nx) from which it emerged—possess conspicuous functions that were not evident in the viruses from which this new clade is alleged to have evolved. These functions include the following:
1) Increased transmissibility leading to markedly faster intercontinental spread.
2) Increased persistence, causing uncharacteristic outbreaks during summer seasons.
3) Increased virulence for both domesticated and wild birds.
4) Increased host range, including a variety of mammal species.
Already in 2017, a team of avian influenza specialists stated that “the emergence and intercontinental spread of highly pathogenic avian influenza A(H5Nx) virus clade 2.3.4.4 is unprecedented.” These researchers noted that the emergence of H5Nx clade 2.3.4.4 is accompanied by a change in receptor-binding specificity, though they also claim that “the potential role of altered receptor specificity in extended host range and the contribution to the rapid worldwide spread of influenza viruses is still unknown.”
According to a 2017 paper by Imai and Kawaoka titled ‘The role of receptor binding specificity in interspecies transmission of influenza viruses,’ receptor binding specificity does indeed play a key role in the interspecies transmission of influenza A viruses. Kawaoka’s research builds on the research published by a R.A.M. Fouchier’s Dutch team in 2010 in a paper titled ‘In Vitro Assessment of Attachment Pattern and Replication Efficiency of H5N1 Influenza A Viruses with Altered Receptor Specificity.’
According to a 2021 paper titled ‘H5Nx Viruses Emerged during the Suppression of H5N1 Virus Populations in Poultry’ by a research team at the University of Georgia:
We show that H5Nx viruses emerged during the successful suppression of H5N1 virus populations in poultry [in China], providing an opportunity for antigenically distinct H5Nx viruses to propagate. Avian influenza vaccination programs would benefit from universal vaccines targeting a wider diversity of influenza viruses to prevent the emergence of novel subtypes.
The findings of these researchers present an illustrative case of Dr. Geert Vanden Bossche’s thesis that mass vaccination with non-sterilizing vaccines can result in the emergence of a new, more virulent viral strain. As the University of Georgia team notes, “In particular, we show that the widespread use of H5N1 vaccines likely conferred a fitness advantage to H5Nx viruses due to the antigenic mismatch of the neuraminidase genes.” From this observation alone, it is evident that human agency (in the form of mass vaccination of poultry in China with non-sterilizing vaccines) contributed to the emergence of the new H5N1 variant.
Currently the world is facing a global pandemic of H5N1 clade 2.3.4.4b—first detected in October 2020 in the Netherlands—that purportedly evolved from H5Nx viruses and possesses even greater pathogenic functions. An especially striking feature of the newly emerged H5N1 clade 2.3.4.4b is how rapidly it spread from birds in Europe to birds in North America. This rapid spread contrasts with the previously slow intercontinental spread of the goose/Guangdong-lineage of H5N1.
After emerging in China in 1996, it was first detected in Europe in 2005, and then in the United States in 2014. While it apparently took nine years for earlier variants to spread from Europe to the United States, H5N1 clade 2.3.4.4b was first detected in the Netherlands October 2020 and then in the United States in late 2021. What could account for the new variant’s extraordinarily rapid intercontinental spread?
In a July 11, 2022 paper in Nature titled ‘Transatlantic spread of highly pathogenic avian influenza H5N1 by wild birds from Europe to North America in 2021,’ a large international team offered the hypothesis that birds migrating from Europe to Iceland and other North Atlantic islands, and from there to North America in 2021, must have carried the virus across the Atlantic. As they noted in their conclusion:
The HPAI H5N1 viruses that were detected in Newfoundland in November and December 2021 originated from Northwest Europe and belonged to HPAI clade 2.3.4.4b. Most likely, these viruses emerged in Northwest Europe in winter 2020/2021, dispersed from Europe in late winter or early spring 2021, and arrived in Newfoundland in autumn 2021. The viruses may have been carried across the Atlantic by migratory birds using different routes, including Icelandic, Greenland/Arctic, or pelagic routes. The unusually high presence of the viruses in European wild bird populations in late winter and spring 2021, as well as the greater involvement of barnacle and greylag geese in the epidemiology of HPAI in Europe since October 2020, may explain why spread to Newfoundland happened this winter (2021/2022), and not in the previous winters.
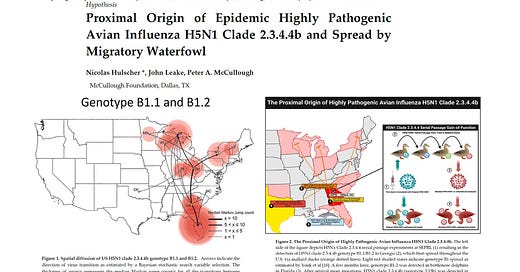
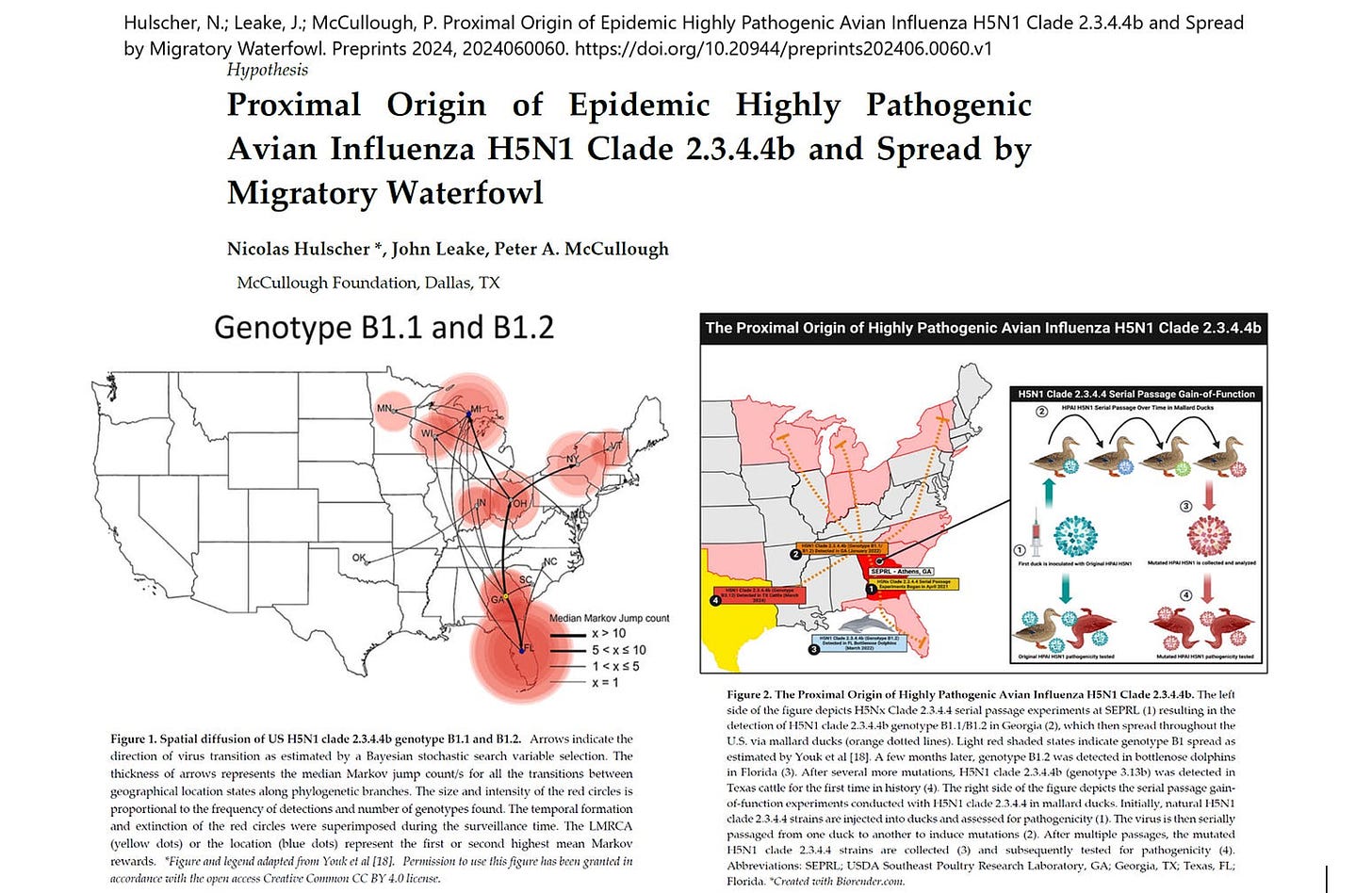
This conclusion contains two implausible elements and a notable omission. First, the hypothetical spread of a new avian influenza variant by migratory birds from Europe to North America by crossing the North Atlantic has never been documented before and therefore appears to be unprecedented. Second, migratory birds in the East Atlantic flyway do not fly from Northwest Europe to North America in the autumn, but the other way around, from North America to Northwest Europe. Third, the paper’s conclusion omits the fact that at the same time (December 2021) the H5N1 clade 2.3.4.4b was purportedly detected in birds in Newfoundland, it was also detected in ducks in South Carolina, just two hundred miles east of the USDA’s Southeast Poultry Research Laboratory (SEPRL), which began performing serial passage experiments with H5Nx viruses on mallard ducks in the spring of 2021. H5Nx viruses share the H5 hemagglutinin (HA) protein but have different neuraminidase (NA) proteins, ranging from H5N1 to H5N9.
HPAI H5N1 belonging to clade 2.3.4.4b (genotype B3.13) are currently infecting a large number and variety of animal species in the United States, resulting in sporadic human infections. In the context of modern viral outbreaks involving pathogenic organisms, it is crucial to evaluate the potential origins of the virus, including the possibility of laboratory involvement. In a recent televised interview, former CDC director Dr. Robert Redfield stated:
In the laboratory, I could make it [H5N1] more infectious to humans in months … it’s been published the four amino acids that I need to change … That’s the real biosecurity threat, that these University labs are doing these bio experiments … Bird Flu, I think, is gonna be the cause of the great pandemic, where they are teaching these viruses how to be more infectious for humans.
The above circumstances prompted us to pose a question—namely, it is possible that HPAI H5N1 clade 2.3.4.4b evolved not in nature, but as a result of serial passage or other Gain-of-Function (GOF) research in a laboratory? The evidence we have uncovered thus far strongly raises the suspicion that this is indeed the case.
—End of Introduction. To read full paper, please click on graphic image below.—
AUTHORS’ NOTE: Because of the huge amount time and effort that went into producing this paper, we contemplated putting it behind a paywall, but then opted to let readers decide if they would like to support our work. If you find it interesting and informative, please become a paid subscriber to our Substack for just $5 per month.
Readers may also consider making a donation to the McCullough Foundation, which is currently planning multiple, high production value audiovisual presentations of our investigative scholarship.
Hulscher, N.; Leake, J.; McCullough, P. Proximal Origin of Epidemic Highly Pathogenic Avian Influenza H5N1 Clade 2.3.4.4b and Spread by Migratory Waterfowl. Preprints 2024, 2024060060. https://doi.org/10.20944/preprints202406.0060.v1
Peter A. McCullough, MD, MPH
President, McCullough Foundation




Because they weren't hung after covid, here we are....
Gates came out last week that been doing “ research “ ( Gain of Function ? ) on strains of bird flu. They being GAVI. Vaccines are already being stockpiled. How would they know which particular human form of H5N1 would be the one to emerge? My guess is they know which one cause they produced it themselves. Of course it’s a mRNA vax.
Just like the COVID pandemic.
CREATE A PROBLEM
SOLVE THE PROBLEM
LOTS of profit to be made 💰💰💰